 |
Jump to: Multiple Alignment Motif Tree Motif Matching
Input file: 10 motifs loaded
Settings: Metric=PCC, Alignment=SWU, Gap-open=1000, Gap-extend=1000, -nooverlapalign
Multiple Alignment=IR, Tree=UPGMA, Matching against: TRANSFAC
Note: All results files are removed nightly at midnight EST. Please save your results by saving "Webpage, complete".
Download results as a PDF
Click here to run STAMP again.
Multiple Alignment
(Consensus sequence representations shown, but multiple alignment was carried out on the matrices)
| DME21: | --AYATTTTT--- |
| DME73: | TTAWCTTT----- |
| DME40: | WAWTATTT----- |
| DME18: | -AWTATTTT---- |
| DME76: | ----TTYTGRAA- |
| DME54: | ----CTCTGKGT- |
| DME39: | -----TKTCTCCT |
| DME64: | -----TTTTTWAT |
| DME48: | -----TTTTCTGA |
| DME15: | -AGGCTTTA---- |
| Familial Profile: (click for matrix) | 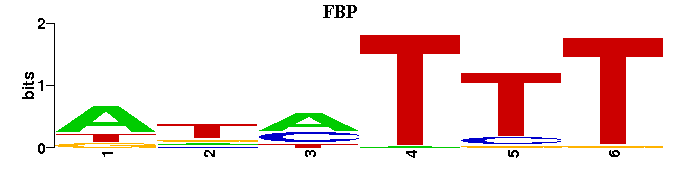 |
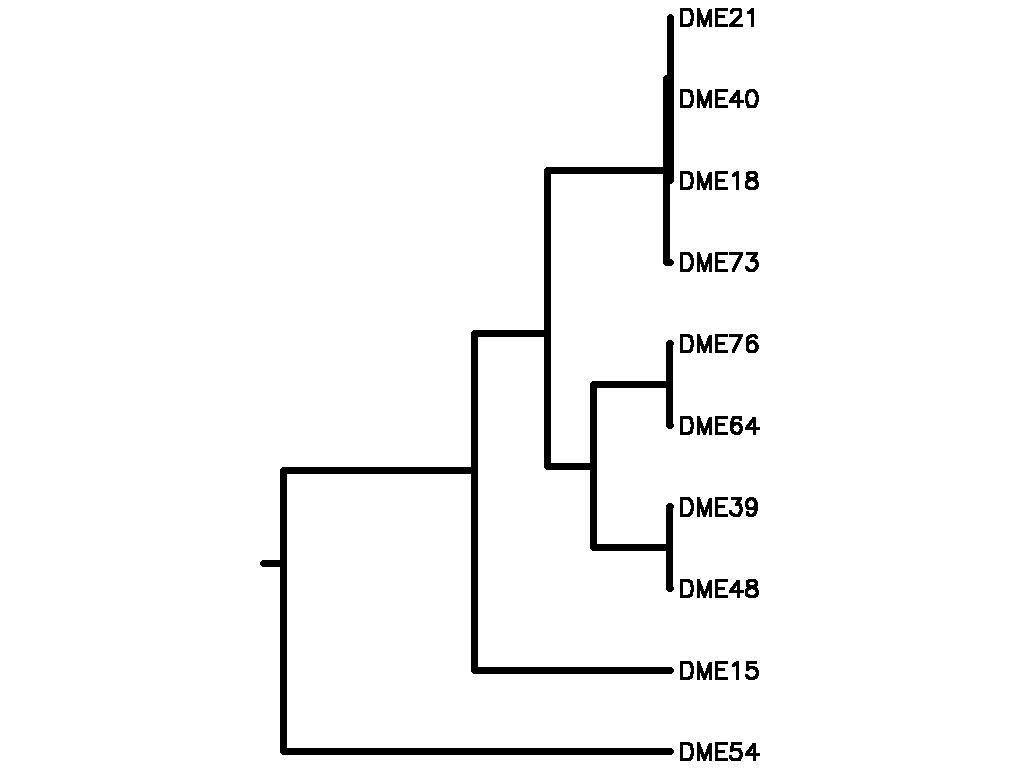
Motif Tree
| Tree (drawn by Phylip) Click here for Newick-format tree (viewable with MEGA) | Input Motif | Best match in TRANSFAC |
 |
|
Motif Similarity Matches
| DME21 |  | 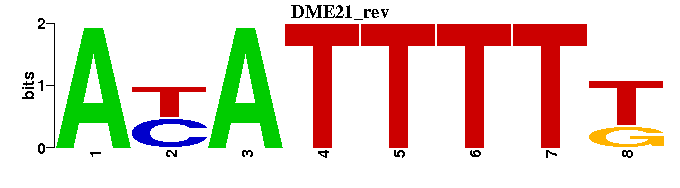 |
| Name | E value | |||
| DMRT7_M01151 | 1.1748e-06 | TGTNACANTKTNG | 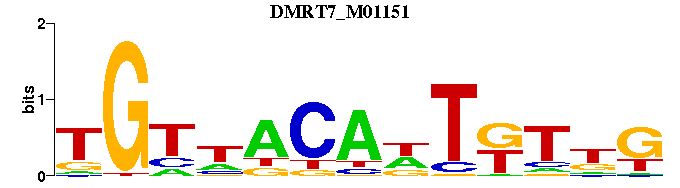 | |
| MCM1+SFF_M01051 | 1.3481e-06 | TNCCNAAWNNGGNAAWANNTAAAYAA |  | |
| FOXJ2_M00423 | 5.1946e-06 | NMAAATATTATKR | 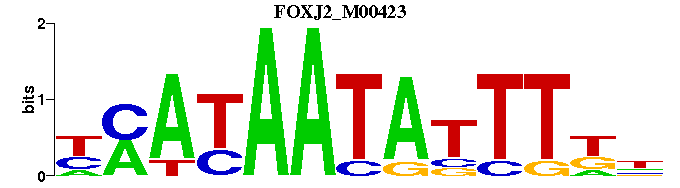 | |
| BR-C_M00092 | 5.3628e-06 | RANAAATAGNWNNNN | 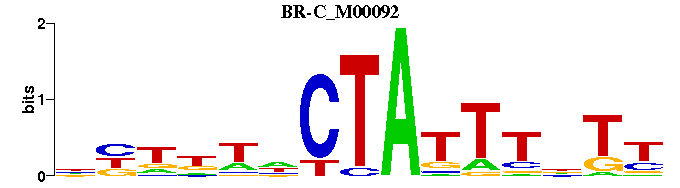 | |
| MEF-2_M00006 | 9.0557e-06 | RGRGTTATTTTTAGA |  |
| DME40 | 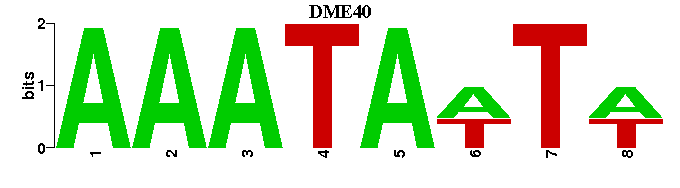 | 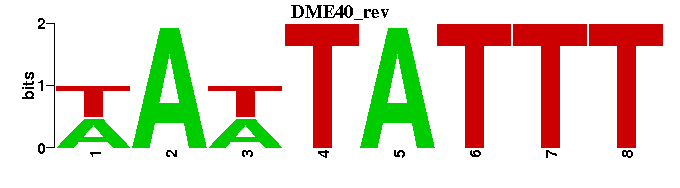 |
| Name | E value | |||
| HNF3beta_M00131 | 2.2341e-08 | GNANTRTTTRYTTW | 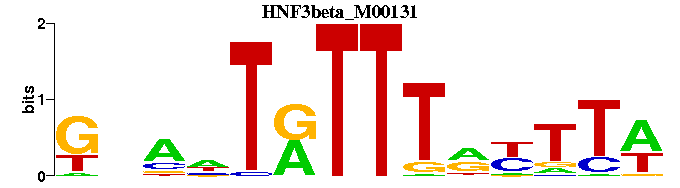 | |
| FOXJ2_M00423 | 4.3076e-07 | YMATAATATTTKN |  | |
| ATHB-5_M00503 | 6.0827e-07 | AATNATTG |  | |
| XFD-1_M00267 | 1.7997e-06 | RYWWTATTTACWT |  | |
| POU1F1_M00744 | 6.3121e-06 | WWTTATNCA | 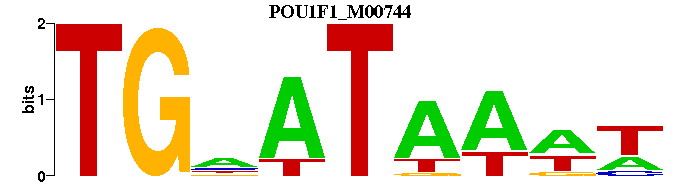 |
| DME18 | 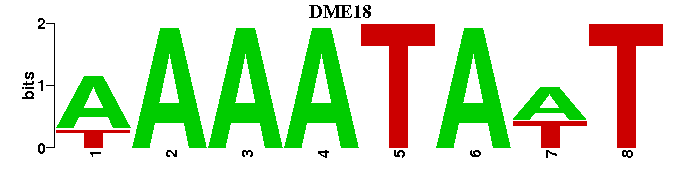 | 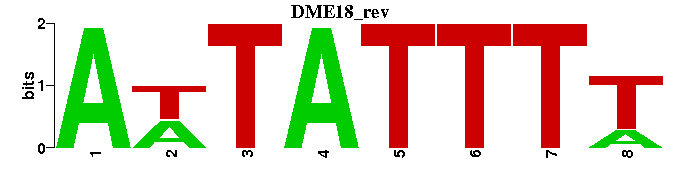 |
| Name | E value | |||
| FOXJ2_M00423 | 4.1244e-08 | NMAAATATTATKR |  | |
| XFD-1_M00267 | 1.4141e-06 | RYWWTATTTACWT |  | |
| MEF-2_M00406 | 2.7500e-06 | KCTAAAAATAACCCN |  | |
| MEF-2_M00006 | 2.9784e-06 | RGRGTTATTTTTAGA |  | |
| Croc_M00266 | 5.2109e-06 | NNNNMTATTTATTNT |  |
| DME73 | 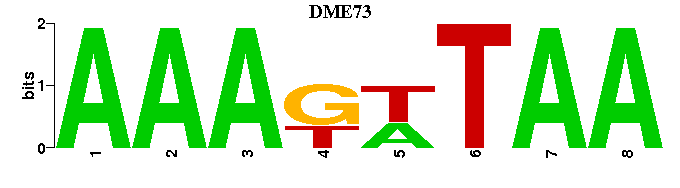 | 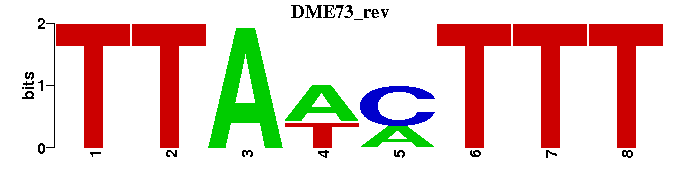 |
| Name | E value | |||
| Evi-1_M00011 | 3.0158e-07 | TTATCTTG |  | |
| Evi-1_M00080 | 3.4111e-06 | TTATCTTATC |  | |
| Evi-1_M00079 | 5.2915e-06 | TTATCTTRTC |  | |
| IRF_M00972 | 5.7982e-06 | AAANWGAAAN | 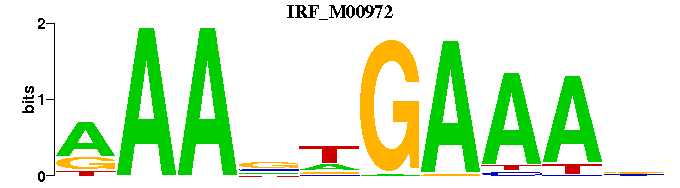 | |
| ISGF-3_M00258 | 1.9424e-05 | GNGAAANWGAAACT | 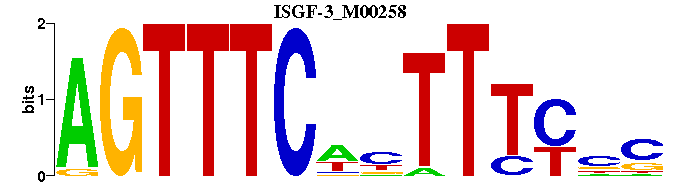 |
| DME76 |  | 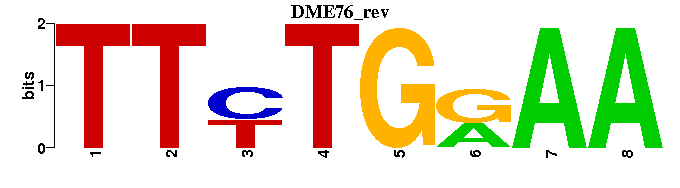 |
| Name | E value | |||
| C-EBPgamma_M00622 | 1.6701e-07 | WTTYTGAAATNA | 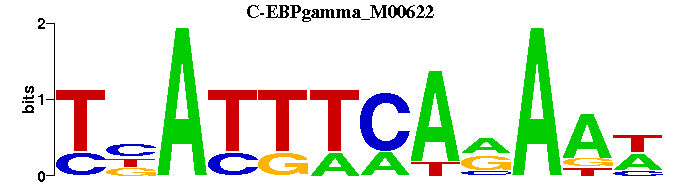 | |
| HSF1_M01023 | 3.4618e-06 | TTCTRGAANNTTCTYM | 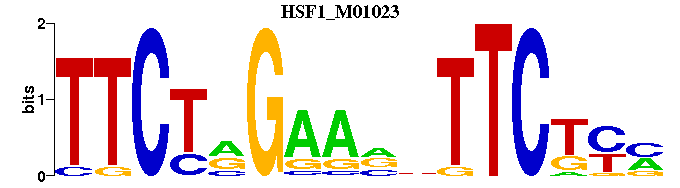 | |
| MADS-A_M00408 | 7.9589e-06 | TTTCCATTTTTNNWN |  | |
| STAT5B_M00459 | 1.5093e-05 | NRNTTCCNGGAAWN | 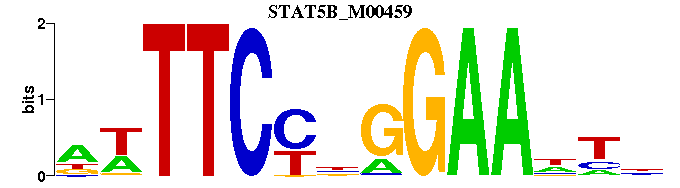 | |
| HSF_M00169 | 1.7698e-05 | NTTCNNGAANNTTC | 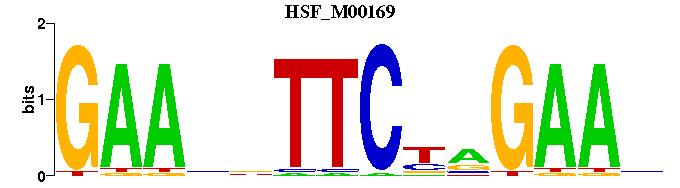 |
| DME64 | 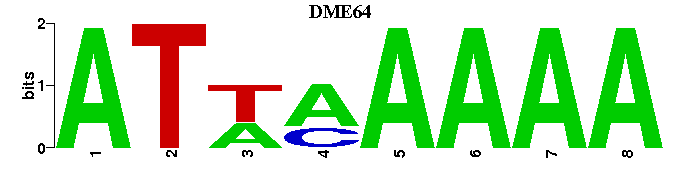 | 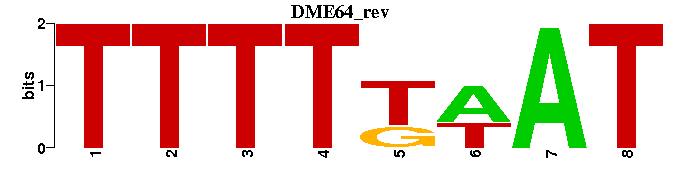 |
| Name | E value | |||
| unc-86_M00689 | 2.1721e-09 | ATTTYKWAT | 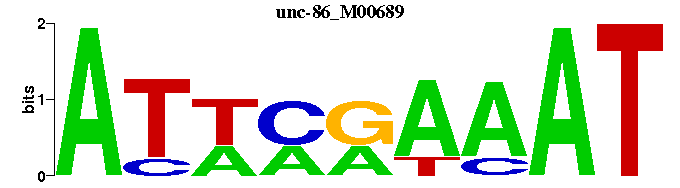 | |
| Hb_M00022 | 2.2334e-06 | NTTTTTNYK | 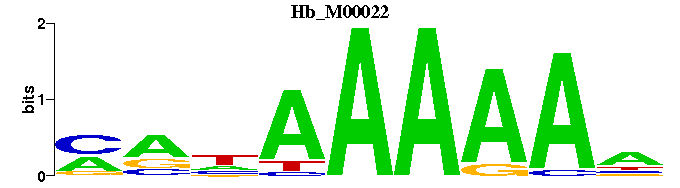 | |
| Nkx6-1_M00424 | 2.8170e-06 | AANCAATTAAAW- |  | |
| FOXP1_M00987 | 4.7303e-06 | ATAAAAAACAACACAAATA | 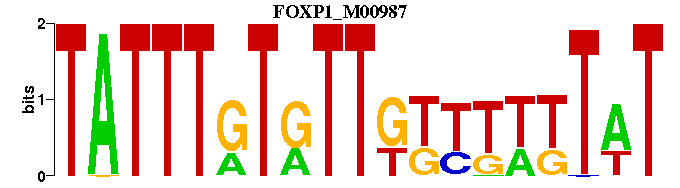 | |
| Abd-B_M01094 | 8.4480e-06 | --TTTTAT |  |
| DME39 | 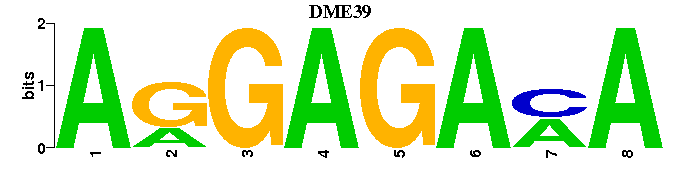 | 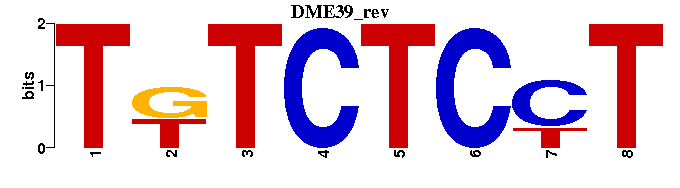 |
| Name | E value | |||
| ARF_M00438 | 4.0015e-06 | --GAGACAA |  | |
| IRF_M00972 | 1.9566e-04 | NTTTCWNTTT |  | |
| ISGF-3_M00258 | 3.6247e-04 | AGTTTCWNTTTCNC |  | |
| HSF_M00163 | 1.1939e-03 | GAANAGAANAGAAN | 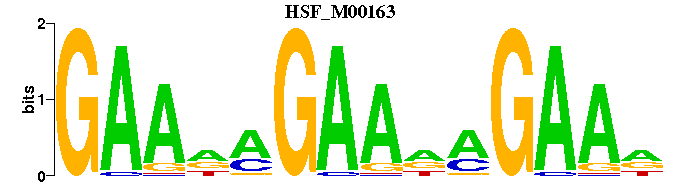 | |
| HSF_M00164 | 1.1939e-03 | GAANAGAANNTTCT | 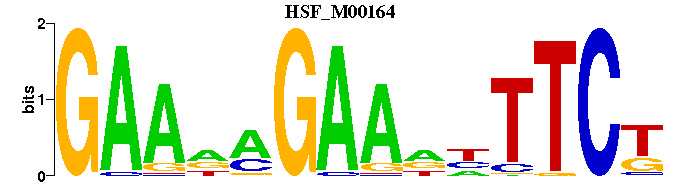 |
| DME48 | 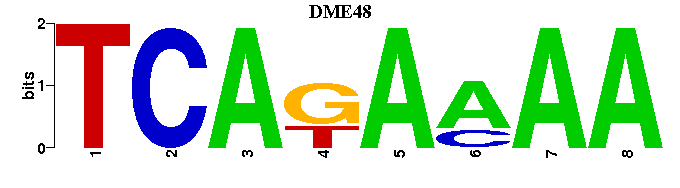 | 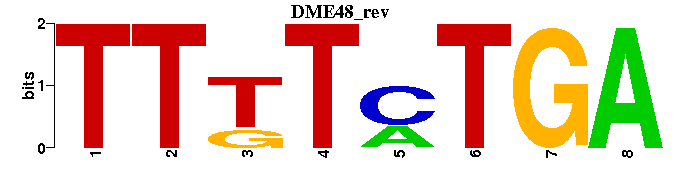 |
| Name | E value | |||
| Abd-B_M01094 | 8.9416e-06 | TTTTAT-- |  | |
| MEIS1A-HOXA9_M00420 | 1.9385e-05 | TCRTAAAMCTGTC |  | |
| MEIS1B-HOXA9_M00421 | 7.6360e-05 | GACASNTTWAYRR |  | |
| Cdx-2_M00729 | 2.5920e-04 | NGYNATAAAANTN | 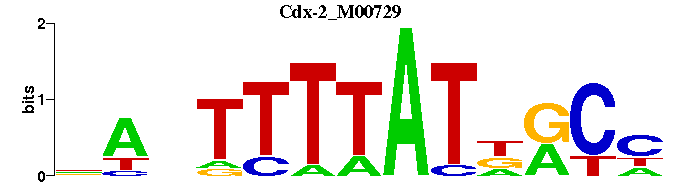 | |
| Hb_M00022 | 2.7174e-04 | -MRNAAAAAN |  |
| DME15 | 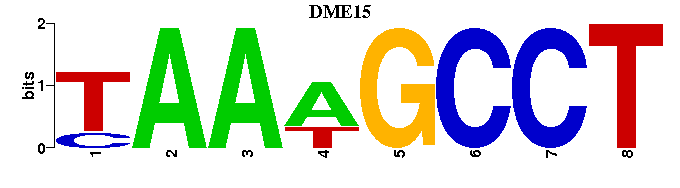 | 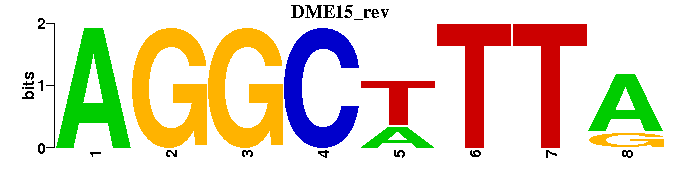 |
| Name | E value | |||
| Retroviral_M00212 | 5.2780e-05 | NAGNRRRRRGNTTTATT |  | |
| c-Rel_M00053 | 5.5959e-04 | GGAAWNYCC | 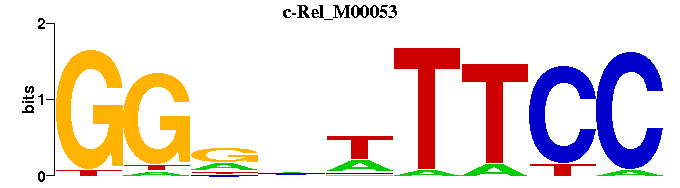 | |
| Lentiviral_M00318 | 5.6138e-04 | ---CTTTATT |  | |
| PLZF_M01075 | 1.2096e-03 | GANCWRNTMAARNTTTANCNNCNRWNNA | 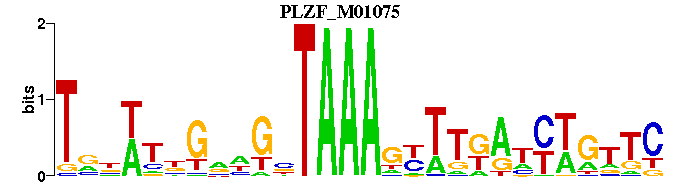 | |
| LEF1_M00805 | 3.5118e-03 | ---CTTTG |  |
| DME54 |  | 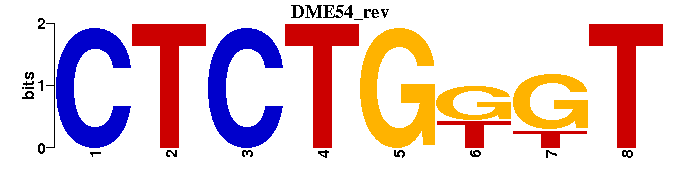 |
| Name | E value | |||
| CP2_M00072 | 3.1033e-04 | --CTGGKTNGNG |  | |
| CACCC-binding_M00721 | 5.3682e-04 | CCNCACCCWNNKGNT | 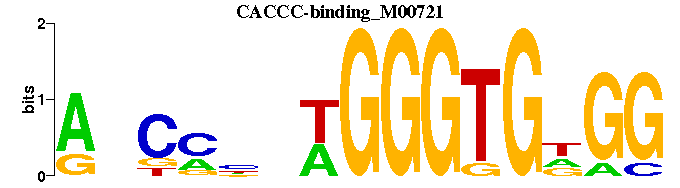 | |
| RAP1_M00213 | 9.8829e-04 | NRTGTATGGGTKY |  | |
| Grainyhead-Elf-1-NTF-1_M00951 | 2.0072e-03 | AAACCRG-- |  | |
| Cdx_M00991 | 2.2904e-03 | TTTATNNNTYTNNYNRT |  |