 |
Jump to: Multiple Alignment Motif Tree Motif Matching
Input file: 10 motifs loaded
Settings: Metric=PCC, Alignment=SWU, Gap-open=1000, Gap-extend=1000, -nooverlapalign
Multiple Alignment=IR, Tree=UPGMA, Matching against: TRANSFAC
Note: All results files are removed nightly at midnight EST. Please save your results by saving "Webpage, complete".
Download results as a PDF
Click here to run STAMP again.
Multiple Alignment
(Consensus sequence representations shown, but multiple alignment was carried out on the matrices)
| Motif1: | TGAWTTCA------ |
| Motif2: | -CANATNTG----- |
| Motif3: | ----TTCTTCAK-- |
| Motif4: | -GTTTGCTS----- |
| Motif5: | TTCCATYY------ |
| Motif6: | ------CTGCAGTG |
| Motif7: | --ATTTCTGT---- |
| Motif8: | ----AGCTSYTK-- |
| Motif9: | -----WCTKTGTT- |
| Motif10: | --CATKSTCT---- |
| Familial Profile: (click for matrix) | 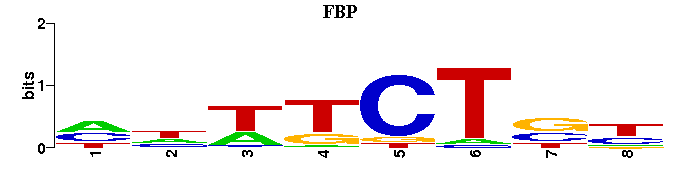 |
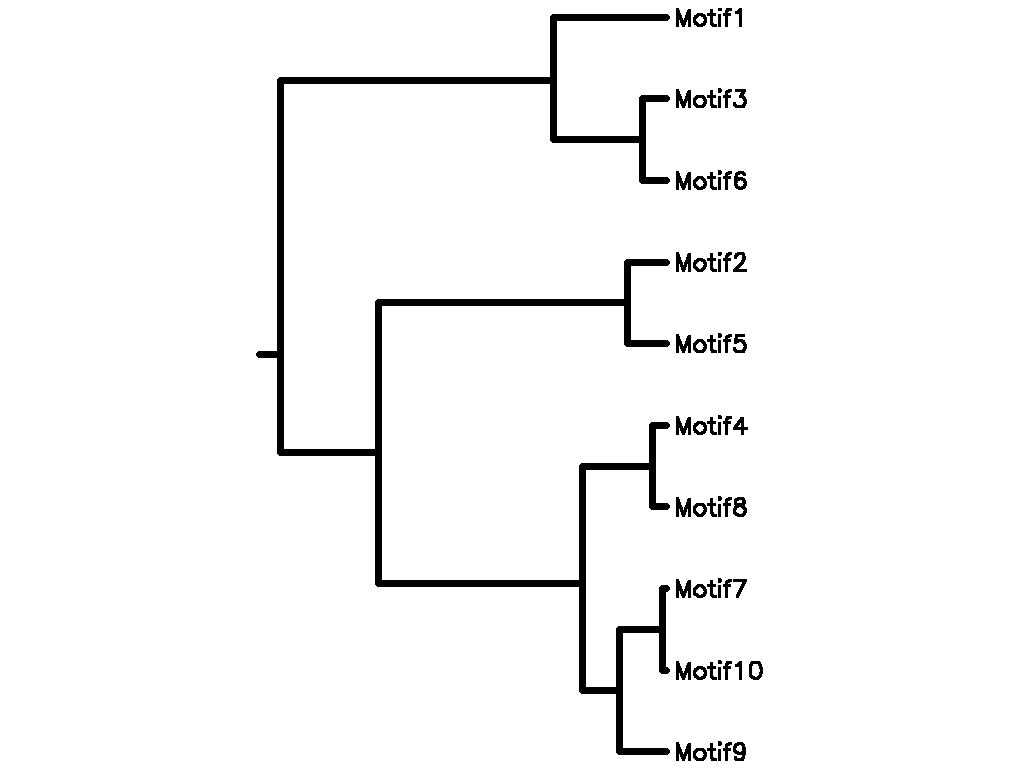
Motif Tree
| Tree (drawn by Phylip) Click here for Newick-format tree (viewable with MEGA) | Input Motif | Best match in TRANSFAC |
 |
|
Motif Similarity Matches
| Motif1 | 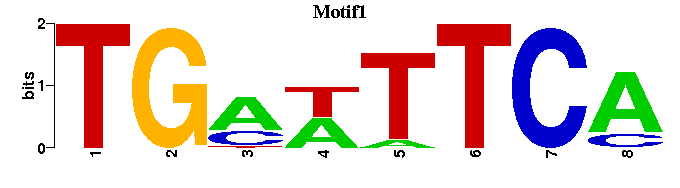 | 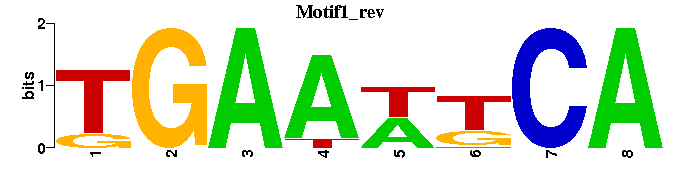 |
| Name | E value | |||
| c-Rel_M00053 | 3.0854e-05 | GGAAWNYCC | 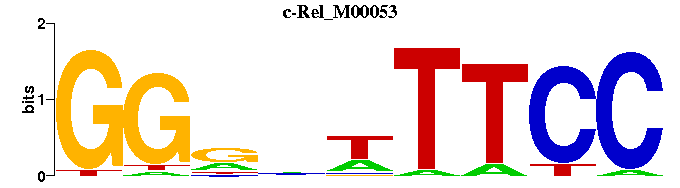 | |
| CF1A_M01098 | 4.9039e-05 | NTTTNAAYTNATTAW | 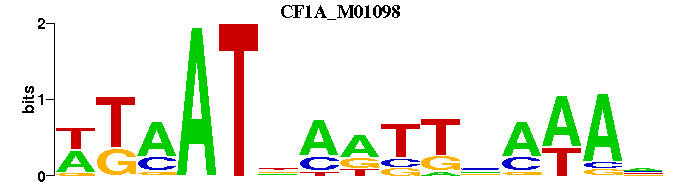 | |
| C-EBPgamma_M00622 | 5.2914e-05 | TNATTTCARAAW | 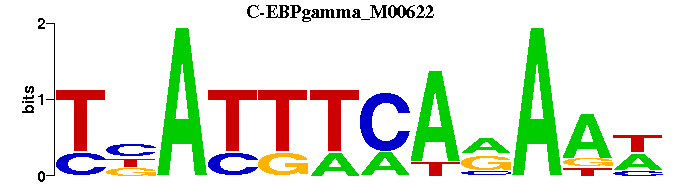 | |
| PXR_M01153 | 2.2677e-04 | TGAACTN- |  | |
| STE12_M00664 | 3.6091e-04 | --GTTTCA |  |
| Motif3 | 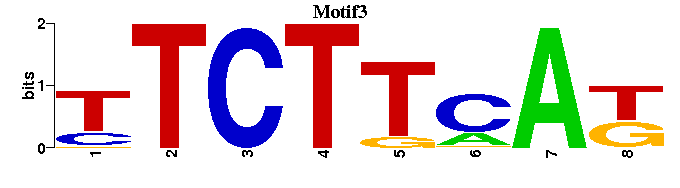 | 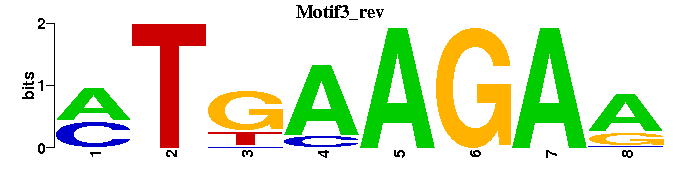 |
| Name | E value | |||
| XPF-1_M00684 | 4.6248e-05 | GKTSNYCNG |  | |
| PEND_M01015 | 2.6082e-04 | ACTTCTT--- |  | |
| POU1F1_M00744 | 3.0913e-04 | WWTTATNCA- | 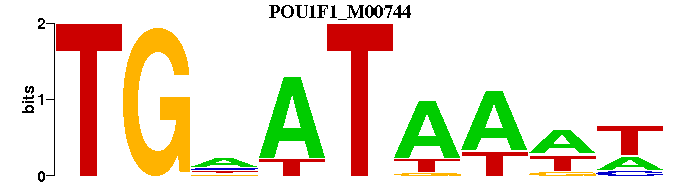 | |
| Tel-2_M00678 | 9.3360e-04 | CAGGAAGTA |  | |
| Nrf-2_M00108 | 1.1291e-03 | SNCTTCCGG |  |
| Motif6 |  | 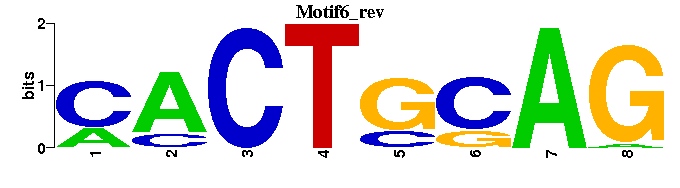 |
| Name | E value | |||
| Gfi1b_M01058 | 2.9222e-04 | -WGCMGTGATTT | 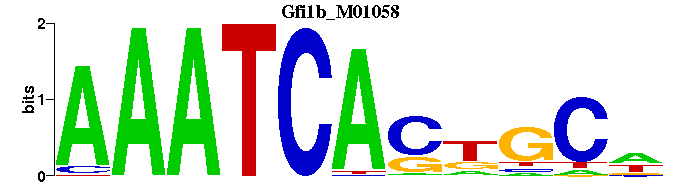 | |
| TTF1_M00432 | 8.0071e-04 | STCAAGTRT |  | |
| Bel-1_M00312 | 1.2384e-03 | ANCTGCTGAYRCWGMGNKNACTNYCNM | 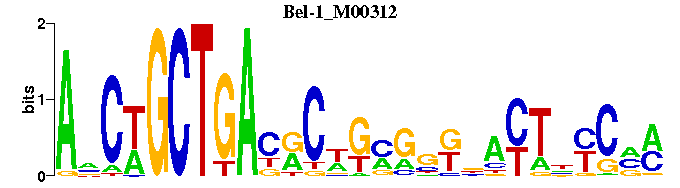 | |
| LXR_M00766 | 2.6279e-03 | GGGTTACTNNCGGTC | 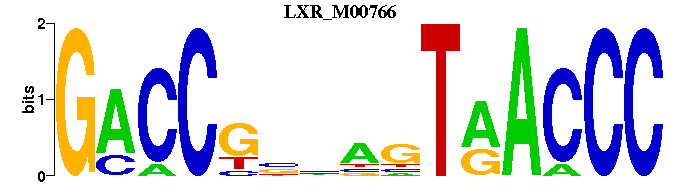 | |
| LXR,_M00965 | 3.1088e-03 | NRGGTYANNNNAGGNC | 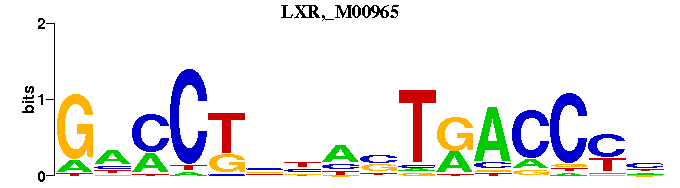 |
| Motif2 | 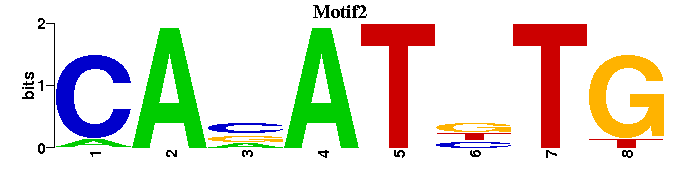 | 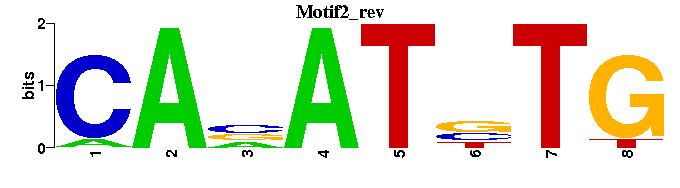 |
| Name | E value | |||
| RP58_M00532 | 1.7629e-05 | TCCAGATGTTN | 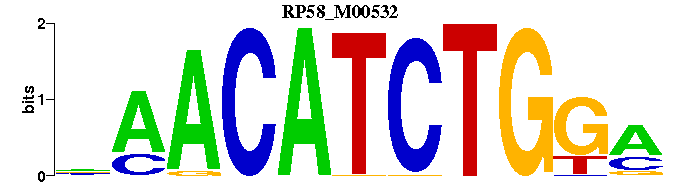 | |
| TFE_M01029 | 3.2230e-05 | NCACATG-- |  | |
| c-Myc-Max_M00123 | 1.2216e-03 | WNNCACGTGNT | 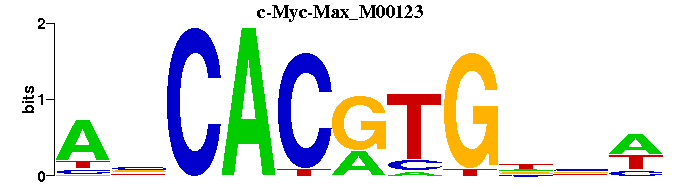 | |
| MyoD_M00001 | 1.7344e-03 | NRNCACCTGNN | 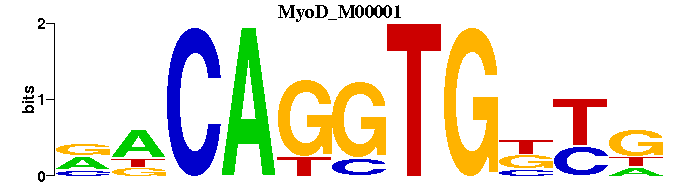 | |
| PHO4_M00064 | 3.7583e-03 | NNCACGTGNGN |  |
| Motif5 | 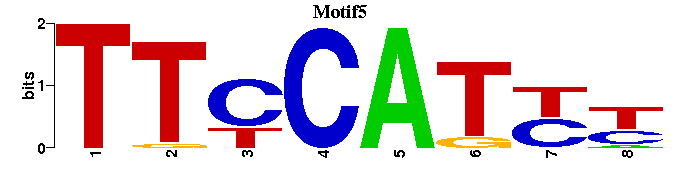 | 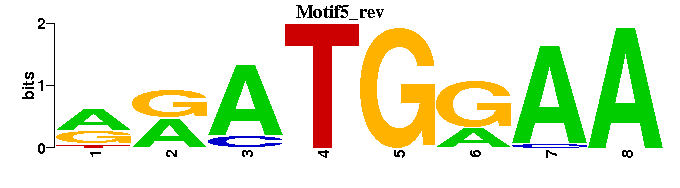 |
| Name | E value | |||
| MADS-A_M00408 | 6.1562e-08 | NWNNAAAAATGGAAA |  | |
| MADS-B_M00404 | 1.8623e-07 | TTTSCATTTTTRGN |  | |
| AG_M01133 | 7.4151e-06 | NTTNCCWTTTTTGNWA | 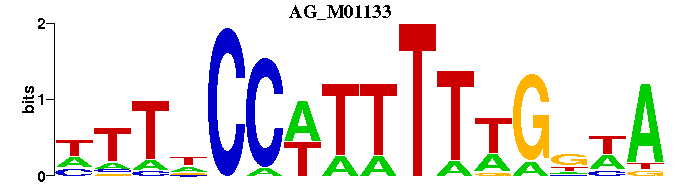 | |
| TEF-1_M00704 | 1.1951e-05 | ---CATYY |  | |
| ICSBP_M00699 | 4.4920e-05 | CAGTTTCAYTT | 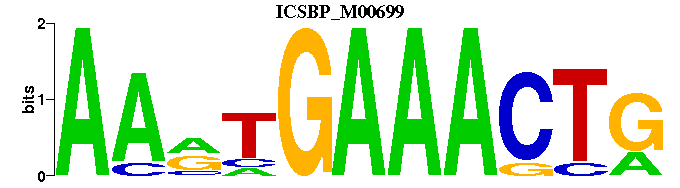 |
| Motif4 | 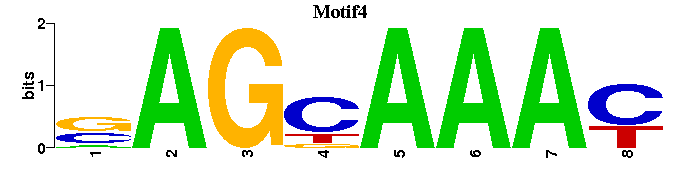 | 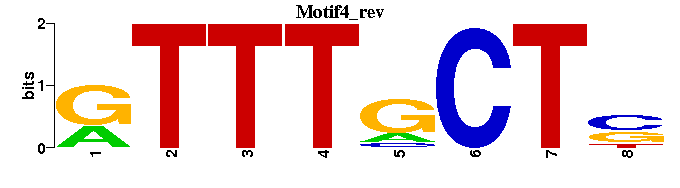 |
| Name | E value | |||
| DBP_M00624 | 1.2145e-06 | --GCANWN | 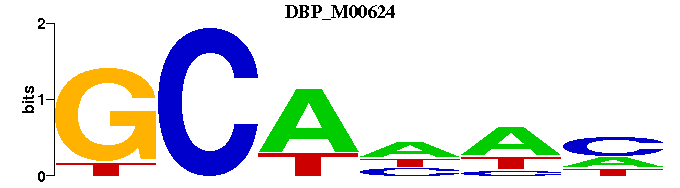 | |
| HNF3alpha_M00724 | 1.7525e-06 | RTTTGYTYWN |  | |
| HNF3_M00791 | 1.6547e-05 | NNTRTTTRYNYN | 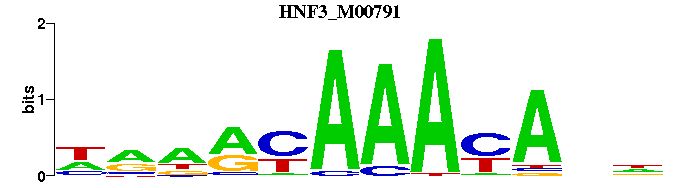 | |
| HNF3beta_M00131 | 9.2151e-05 | WAARYAAAYANTNC | 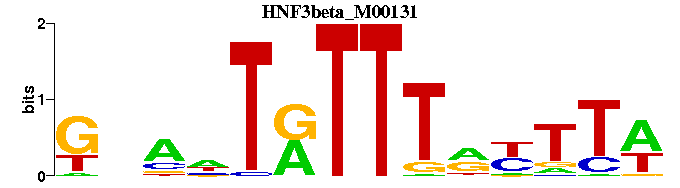 | |
| dl_M00120 | 9.5071e-05 | NGNTTTTCYC |  |
| Motif8 | 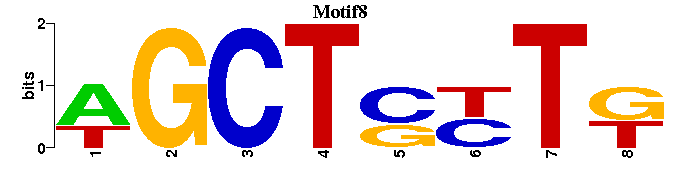 | 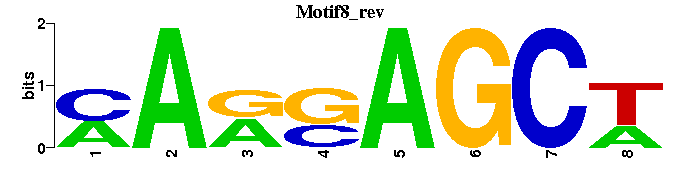 |
| Name | E value | |||
| AP-4_M00005 | 5.1179e-04 | NNNNCAGCTGNNGNCNK |  | |
| AP-4_M00927 | 5.2612e-04 | --RCAGCTGN |  | |
| TAL1_M00993 | 1.1218e-03 | -RNCAGNTGG |  | |
| Barbie_M00238 | 1.3703e-03 | NNNAAAGSNGNNKG | 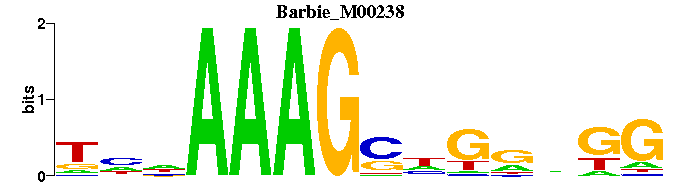 | |
| ZBRK1_M01105 | 1.4964e-03 | AAANNNCTGCKSCC |  |
| Motif7 | 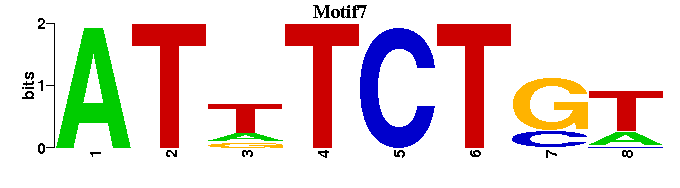 | 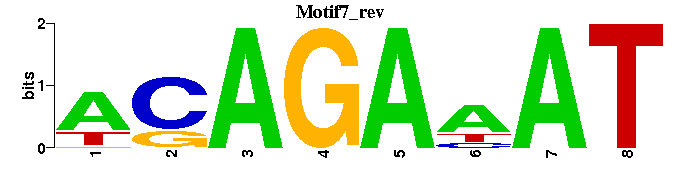 |
| Name | E value | |||
| SEF-1_M00214 | 5.7846e-06 | ACACGGATATCTGTGGTY |  | |
| STAT1_M00492 | 7.2644e-05 | -SGGAANT |  | |
| PEND_M01015 | 1.9853e-04 | ACTTCTT- |  | |
| FOXP1_M00987 | 1.9933e-04 | ATAAAAAACAACACAAATA | 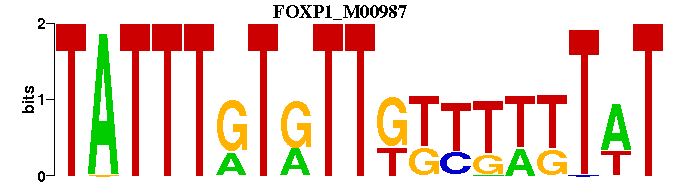 | |
| HMG_M00750 | 3.7691e-04 | AWTTTC--- |  |
| Motif10 | 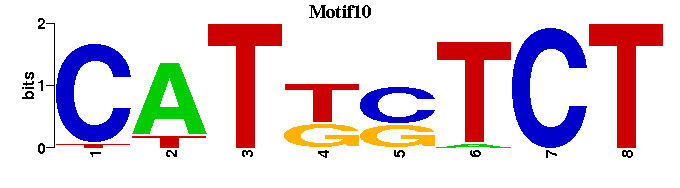 | 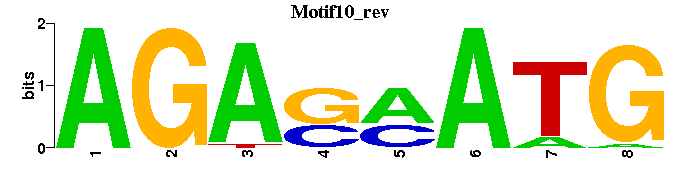 |
| Name | E value | |||
| Mat1-Mc_M00276 | 1.4858e-05 | CNATTGTYN |  | |
| alpha-CP1_M00687 | 8.4603e-05 | AGCCAATGAG |  | |
| ARF_M00438 | 1.2164e-04 | GAGACAA-- |  | |
| Alx-4_M00619 | 5.5772e-04 | GATTATTCTCAG |  | |
| SOX17_M01016 | 1.0310e-03 | -ATTGT-- |  |
| Motif9 | 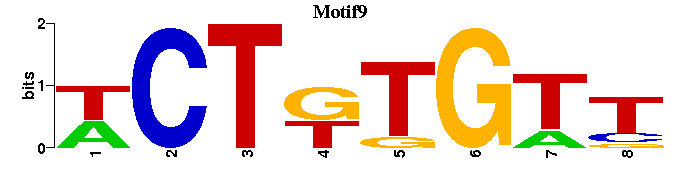 | 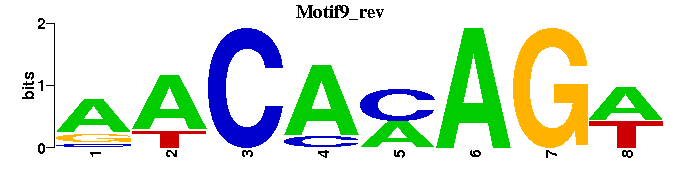 |
| Name | E value | |||
| STE11_M01005 | 1.4657e-08 | TTTCTTTGTTC |  | |
| SOX_M01014 | 6.3476e-08 | TCTTTGTTANGN |  | |
| STE11_M00274 | 3.2681e-07 | WNNNNAACAAAGAAANW | 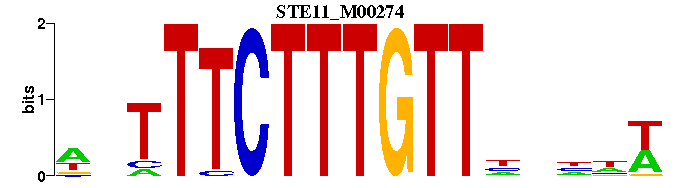 | |
| Mat1-Mc_M00275 | 5.1737e-07 | NRMNNAACAAAGANRYC |  | |
| SRY_M00148 | 7.5859e-06 | --KNTGTT |  |